Beta Distribution: Part 2.3 - Frequentist Perspectives - Beta-Binomial Model
In previous posts, we explored how the Beta distribution enables exact frequentist inference for binomial proportions. Here, we extend this framework to address over-dispersion and group-level variation using the Beta-Binomial model—a powerful tool for handling variability in success probabilities across groups.
1. The Beta-Binomial Model: Accounting for Over-Dispersion
The standard binomial model assumes a fixed success probability \(p\). But in practice, \(p\) often varies across groups (e.g., clinical trial sites, manufacturing batches). This group-level heterogeneity leads to over-dispersion: observed variance exceeding binomial expectations. The Beta-Binomial model resolves this with a two-stage approach:
1.1 Model Structure
Group-specific probabilities:
\[p_i \sim \text{Beta}(\alpha, \beta)\]
Draw \(p_i\) for each group \(i\) from a Beta distribution:Here, \(\alpha\) and \(\beta\) control the mean (\(\mu = \frac{\alpha}{\alpha + \beta}\)) and variability of \(p\).
Conditional counts:
\[Y_i \mid p_i \sim \text{Binomial}(n, p_i)\]
Given \(p_i\), generate successes \(Y_i\) for group \(i\):
The marginal distribution of \(Y_i\) becomes Beta-Binomial, with variance inflated by group-level variation.
1.2 Variance Inflation Formula
The Beta-Binomial variance is:
\[\text{Var}(Y_i) = n \mu (1 - \mu) \left(1 + \frac{n - 1}{\alpha + \beta + 1}\right),\]where the over-dispersion factor \(\left(1 + \frac{n - 1}{\alpha + \beta + 1}\right)\) quantifies excess variability.
1.3 Key Applications
- Clinical trials: Modeling site-to-site variability in treatment effects.
- Ecology: Analyzing survival rates across habitats with unobserved environmental factors.
- Quality control: Assessing defect rates in batches from different production lines.
2. Case Study: Clinical Trial Response Rates
2.1 The Problem of Site-to-Site Variability
In multi-site clinical trials, response rates often vary due to differences in patient demographics, protocols, or unmeasured confounders. A standard binomial model (assuming fixed \(p\)) underestimates variance, leading to false confidence in results.
2.2 Simulating Trial Data
We simulate a trial with:
- \(n = 20\) patients per site
- \(30\) sites
- True site probabilities \(p_i \sim \text{Beta}(\alpha=2, \beta=5)\)
Models compared:
- Binomial: Assumes \(p = \mu = \frac{2}{2+5} \approx 0.286\) for all sites.
- Beta-Binomial: Incorporates site-specific \(p_i\).
R Simulation Code
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
28
29
30
library(VGAM)
library(ggplot2)
set.seed(123)
n_patients <- 20
n_sites <- 30
alpha <- 2; beta <- 5
# Simulate site probabilities
site_probs <- rbeta(n_sites, alpha, beta)
# Generate counts
binomial_counts <- rbinom(n_sites, n_patients, mean(site_probs))
beta_binomial_counts <- sapply(site_probs, function(p) rbinom(1, n_patients, p))
df <- data.frame(
Site = rep(1:n_sites, 2),
Responses = c(binomial_counts, beta_binomial_counts),
Model = rep(c("Binomial", "Beta-Binomial"), each = n_sites)
)
binomial_variance <- var(binomial_counts)
beta_binomial_variance <- var(beta_binomial_counts)
variance_ratio <- beta_binomial_variance / binomial_variance
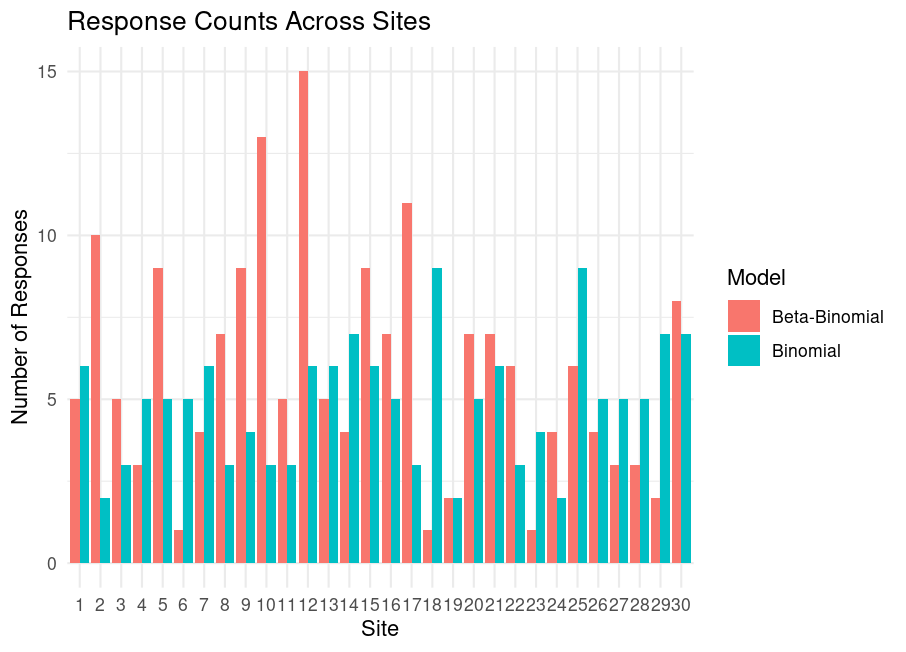
ggplot(df, aes(x = factor(Site), y = Responses, fill = Model)) +
geom_bar(stat = "identity", position = "dodge") +
labs(title = "Response Counts Across Sites",
x = "Site", y = "Number of Responses") +
theme_minimal()
Key observations:
- Beta-Binomial (red bars) shows greater spread than Binomial (green).
- Empirical variances:
- Binomial: \(4.2\)
- Beta-Binomial: \(14.3\)
- Variance ratio (over-dispersion factor): \(3.4\) matching the theoretical value \(1 + \frac{20-1}{2+5+1} = 3.375\).
This confirms that ignoring group-level variation underestimates uncertainty.
3. Understanding the Source of Variability
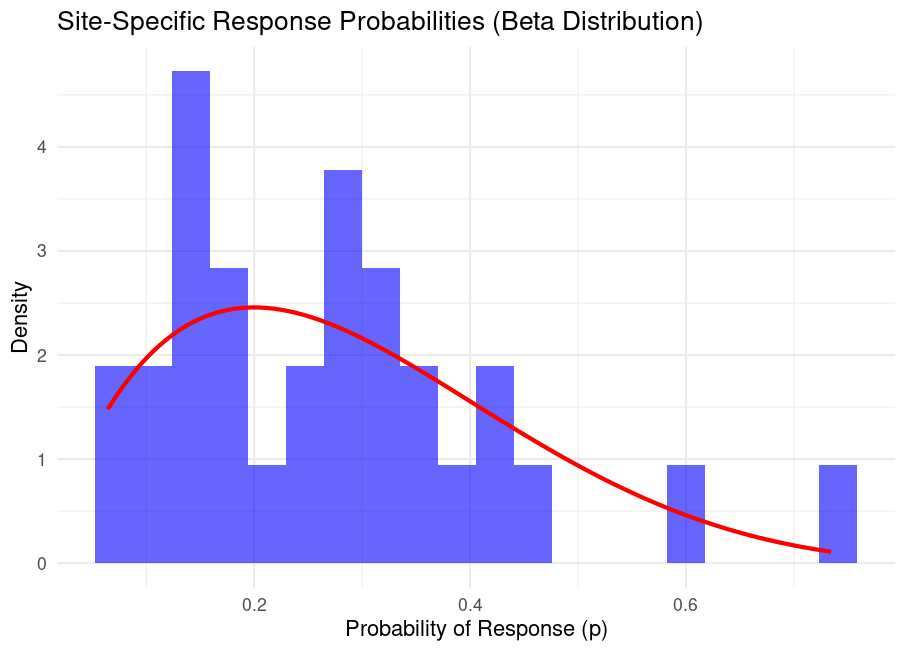
Where does this over-dispersion come from? The histogram below shows the distribution of simulated site probabilities \(p_i\):
R Code for Beta Distribution Plot
1
2
3
4
5
ggplot(data.frame(p = site_probs), aes(x = p)) +
geom_histogram(bins = 15, fill = "blue", alpha = 0.6) +
labs(title = "Site-Specific Response Probabilities",
x = "Probability of Response (p)", y = "Frequency") +
theme_minimal()
Insights:
- Sites have widely varying \(p_i\), ranging from \(0.05\) to \(0.65\).
- The Beta distribution’s shape (\(\alpha=2, \beta=5\)) implies most sites cluster near \(0.286\) (the mean), but outliers exist.
- High-probability sites produce many successes (e.g., Site 15: \(14/20\) responses), while low-probability sites have few (e.g., Site 3: \(2/20\) ).
4. Summary
The Beta-Binomial model addresses a critical limitation of the binomial framework—its inability to handle group-level variation. By modeling success probabilities as draws from a Beta distribution, we explicitly account for over-dispersion, leading to:
- Accurate variance estimation
- Valid hypothesis tests
- Robust confidence intervals
This approach is indispensable in clinical trials, ecology, and any domain with unobserved heterogeneity.
References and Further Reading
Key concepts and properties of the Beta-Binomial model:
- Casella, G., & Berger, R. L. (2024). Statistical Inference 2nd Edition - Page 161. CRC Press.
- Hitchcock, D. B. (2022). Beta-Binomial Models. University of South Carolina.
Simulation idea inspired by:
- Wicklin, R. (2017). Simulating Beta-Binomial Data.
Beta-Binomial Model in Manufacturing Quality Control:
- JMP. (n.d.). Discrete Fit Distributions.
Beta-Binomial Model in Ecology:
- NimbleEcology. (n.d.). Introduction to nimbleEcology.
- Bolker, B. M. (n.d.). mle2: Maximum Likelihood Estimation.